Moisture Advection
import numpy as np
import matplotlib.pyplot as plt
import metpy.calc as mcalc
from metpy.units import units
import glob
import xarray as xr
from datetime import datetime, timedelta
from functools import partial
import numpy as np
import pandas as pd
import xarray as xr
import matplotlib
import matplotlib.animation as animation
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from matplotlib.colors import Normalize
path_staging = "/data/project/ARM_Summer_School_2024_Data/lasso_tutorial/cacti/lasso-cacti"
file_list = sorted(glob.glob(f'{path_staging}/20190129/eda09/base/les/subset_d3/corlasso_met_*'))
ds = xr.open_dataset(file_list[30])
ds
<xarray.Dataset> Size: 6GB
Dimensions: (Time: 1, south_north: 865, west_east: 750,
bottom_top: 149)
Coordinates:
* Time (Time) datetime64[ns] 8B 2019-01-29T13:30:00
XLONG (south_north, west_east) float32 3MB ...
XLAT (south_north, west_east) float32 3MB ...
XTIME (Time) float32 4B ...
Dimensions without coordinates: south_north, west_east, bottom_top
Data variables: (12/52)
ITIMESTEP (Time) int32 4B ...
MUTOT (Time, south_north, west_east) float32 3MB ...
HGT (Time, south_north, west_east) float32 3MB ...
HAMSL (Time, bottom_top, south_north, west_east) float32 387MB ...
P_HYD (Time, bottom_top, south_north, west_east) float32 387MB ...
PRESSURE (Time, bottom_top, south_north, west_east) float32 387MB ...
... ...
MULFC (Time, south_north, west_east) float32 3MB ...
MULNB (Time, south_north, west_east) float32 3MB ...
MULPL (Time, south_north, west_east) float32 3MB ...
MUCAPE (Time, south_north, west_east) float32 3MB ...
MUCIN (Time, south_north, west_east) float32 3MB ...
REFL_10CM_MAX (Time, south_north, west_east) float32 3MB ...
Attributes: (12/39)
DX: 500.0
DY: 500.0
SIMULATION_START_DATE: 2019-01-29_06:00:00
WEST-EAST_GRID_DIMENSION: 751
SOUTH-NORTH_GRID_DIMENSION: 866
BOTTOM-TOP_GRID_DIMENSION: 150
... ...
doi_isPartOf_lasso-cacti: https://doi.org/10.5439/1905789
doi_isDocumentedBy: https://doi.org/10.2172/1905845
doi_thisFileType: https://doi.org/10.5439/1905819
history: processed by user d3m088 on machine cirrus89...
filename_user: corlasso_met_2019012900eda09d3_base_M1.m1.20...
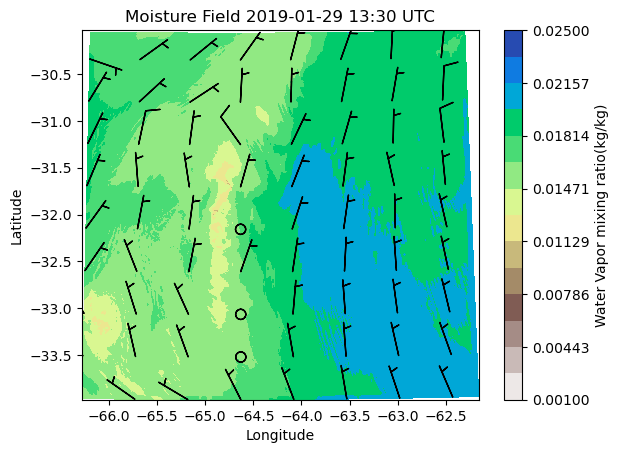
filename_storage: corlassomet2019012900eda09d3baseM1.m1.201901...theta = ds.QVAPOR[0,0,:,:]
u = ds.UMET10[0,:,:]
v = ds.VMET10[0,:,:]
space =100
cf=plt.contourf(ds.XLONG,ds.XLAT,theta, cmap='terrain_r', levels=np.linspace(0.001,0.025, 15))
plt.xlabel('Longitude')
plt.ylabel('Latitude')
time_label=plt.title('Moisture Field')
tx_time= plt.title(f"Moisture Field {pd.to_datetime(ds['Time'])[0]:%Y-%m-%d %H:%M} UTC")
ba = plt.barbs(ds.XLONG [::space, ::space], ds.XLAT[::space, ::space], u[::space, ::space],v[::space, ::space])
plt.colorbar(cf, label='Water Vapor mixing ratio(kg/kg)')
<matplotlib.colorbar.Colorbar at 0x7f8eae6e0e90>
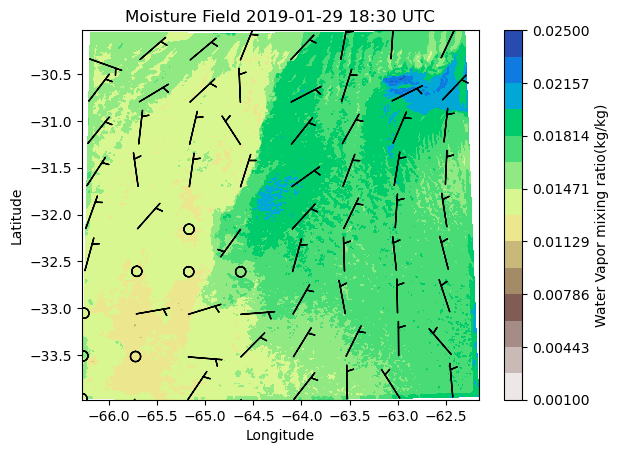
ds = xr.open_dataset(file_list[50])
theta = ds.QVAPOR[0,0,:,:]
u = ds.UMET10[0,:,:]
v = ds.VMET10[0,:,:]
space =100
cf=plt.contourf(ds.XLONG,ds.XLAT,theta, cmap='terrain_r', levels=np.linspace(0.001,0.025, 15))
plt.xlabel('Longitude')
plt.ylabel('Latitude')
time_label=plt.title('Moisture Field')
tx_time= plt.title(f"Moisture Field {pd.to_datetime(ds['Time'])[0]:%Y-%m-%d %H:%M} UTC")
ba = plt.barbs(ds.XLONG [::space, ::space], ds.XLAT[::space, ::space], u[::space, ::space],v[::space, ::space])
plt.colorbar(cf, label='Water Vapor mixing ratio(kg/kg)')
<matplotlib.colorbar.Colorbar at 0x7f8eae500e90>
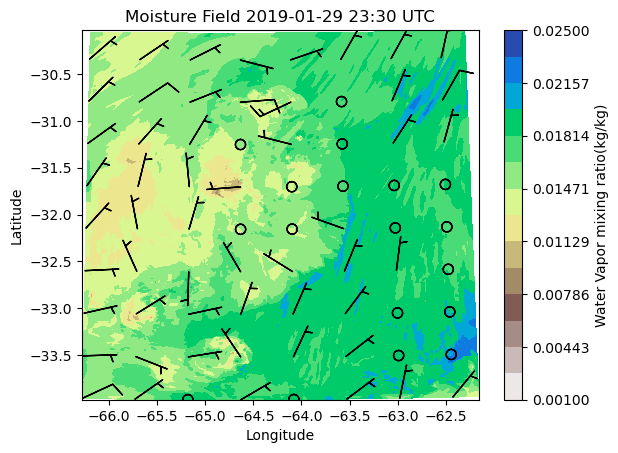
ds = xr.open_dataset(file_list[70])
theta = ds.QVAPOR[0,0,:,:]
u = ds.UMET10[0,:,:]
v = ds.VMET10[0,:,:]
space =100
cf=plt.contourf(ds.XLONG,ds.XLAT,theta, cmap='terrain_r', levels=np.linspace(0.001,0.025, 15))
plt.xlabel('Longitude')
plt.ylabel('Latitude')
time_label=plt.title('Moisture Field')
tx_time= plt.title(f"Moisture Field {pd.to_datetime(ds['Time'])[0]:%Y-%m-%d %H:%M} UTC")
ba = plt.barbs(ds.XLONG [::space, ::space], ds.XLAT[::space, ::space], u[::space, ::space],v[::space, ::space])
plt.colorbar(cf, label='Water Vapor mixing ratio(kg/kg)')
<matplotlib.colorbar.Colorbar at 0x7f8eae3d7710>
Cross Sections
path_staging = "/data/project/ARM_Summer_School_2024_Data/lasso_tutorial/cacti/lasso-cacti"
file_list = sorted(glob.glob(f'{path_staging}/20190129/eda09/base/les/subset_d3/corlasso_met_*'))
ntimes=73
ds= xr.open_mfdataset(file_list[:ntimes])
file_list = sorted(glob.glob(f'{path_staging}/20190129/eda09/base/les/subset_d3/corlasso_cld_*'))
ds2= xr.open_mfdataset(file_list[:ntimes])
# User settings...
# Case date and times to plot:
case_date = datetime(2019, 1, 29)
single_time = case_date + timedelta(hours=20, minutes=15) # time for single-time plots
# Simulation selection within the case date:
ens_name = "eda09"
config_label = "base"
domain = 2
plot_lat= -32.1264 #S
# Next, we have to figure out which j-index to sample for the desired latitude. WRF's coordinate name for this dimension is south_north.
# We will ignore the map projection and pull along a single j index, which we will determine from the middle of the grid.
lats = ds2["XLAT"]
lons = ds2["XLONG"]
nlat, nlon = lons.shape
abslat = np.abs(lats.isel(west_east=int(nlon/2)) - plot_lat)
jloc = np.argmin(abslat.values)
lonslice = lons.isel(south_north=jloc)
hamsl_raw = ds['HAMSL'].isel(Time=0, south_north=jloc) # height of each grid cell on the raw model levels
# ...and now the data we actually want to plot, which is clouds
plotdata_raw = 1000.*(
ds2['QCLOUD'].sel(south_north=jloc) +
ds2['QICE'].sel(south_north=jloc)
) # cloud water+ice mixing ratio converted go g/kg
#pcm=plotdata_raw.isel(Time=0).plot()
def plot_subsequent_frame(itime, plotdata_raw, pcm, tx_time, ba, ba_U, ba_V,ifreq):
"""
Subroutine to plot the next frame of an animation after the first frame is already defined.
This is to be called by animation.FuncAnimation.
Note that some data is pulled from global scope. Variables accessed from outside the subroutine:
pcm = pcolormesh object from the initial frame
tx_time = text annotation object for the time label from the initial frame
plotdata_raw = variable holding the time series of data to plot
plot_lat = latitude being plotted (needed to reconstruct the title text)
"""
print("plotting frame ", itime)
# Overwrite the data in memory for the values displayed in the plot...
# NOTE: This is technically a faux paux for WRF since the model levels are not constant in time. BE CAREFUL!
# However, in this situation the levels are not going to change so much as to make the plot visibly change much.
pcm.set_array(plotdata_raw.isel(Time=itime).values.ravel())
print(ba_U.isel(Time=itime).values[::ifreq, ::ifreq].shape)
print(ba_V.isel(Time=itime).values[::ifreq, ::ifreq].shape)
#ba.set_UVC(ba_U.isel(Time=itime).values[::ifreq, ::ifreq].ravel(),ba_V.isel(Time=itime).values[::ifreq, ::ifreq].ravel())
# Update the time label...
time_label = f"{pd.to_datetime(plotdata_raw['Time'])[itime]:%Y-%m-%d %H:%M} UTC"
tx_time.set_text(f"Cloud Condensate; \n{time_label}")
# end plot_subsequent_frame()
fig, ax = plt.subplots(1, 1, figsize=[8,3.5])
pcm= ax.pcolormesh(lonslice.values, hamsl_raw.values, plotdata_raw.isel(Time=0).values,
shading='auto',norm=Normalize(vmin=0,vmax=1.25))
#pcm=plotdata_raw.plot()
#theta = ds.QVAPOR[0,0,:,:]
da_u = ds["UMET"].isel(south_north=jloc)
da_v = ds["VMET"].isel(south_north=jloc)
space =100
cf=plt.contourf(ds.XLONG,ds.XLAT,theta)
#plt.xlabel('Longitude')
#plt.ylabel('Latitude')
tx_time= plt.title(f"Moisture Field {pd.to_datetime(plotdata_raw['Time'])[0]:%Y-%m-%d %H:%M} UTC")
lonslice2d = (lonslice+(hamsl_raw*0)).T
print(lonslice2d.T.shape)
#print(hamsl_raw.isel(Time=0).shape)
#print(da_u.isel(Time=0).shape)
da_u = ds["UMET"].isel(south_north=jloc)
da_v = ds["VMET"].isel(south_north=jloc)
ifreq = 10
print(lonslice2d.values[::ifreq,::ifreq].shape)
print(hamsl_raw.values[::ifreq,::ifreq].shape)
print(da_u.isel(Time=0).values[::ifreq,::ifreq].shape)
print(da_v.isel(Time=0).values[::ifreq,::ifreq].shape)
#plt.barbs(lonslice2d.values[::ifreq,::ifreq], hamsl_raw.values[::ifreq,::ifreq], da_u.isel(Time=0).values[::ifreq,::ifreq], da_v.isel(Time=0).values[::ifreq,::ifreq],color='w')
# plt.pcolormesh(lonslice.values, hamsl_raw.isel(Time=0).values, da_v.isel(Time=0).values)
plt.show()
plt.colorbar(cf, label='Water Vapor mixing ratio(kg/kg)')
#-----------------------------------------------------------------------
# Now, update the underlying data in the plot for each subsequent frame...
# NOTE: See https://matplotlib.org/stable/api/_as_gen/matplotlib.animation.FuncAnimation.html for
# explaination of how to use functools.partial for passing arguments to the redraw subroutine.
ani = animation.FuncAnimation(fig,
partial(plot_subsequent_frame, plotdata_raw=plotdata_raw, pcm=pcm, tx_time=tx_time,ba=ba,ba_U=da_u,ba_V=da_v,ifreq=ifreq),
frames=range(ntimes))
# And save the results...
filename_mp4 = f"./corlasso_anim_qci_{case_date:%Y%m%d}00{ens_name}d{domain}_{config_label}.mp4"
fps = 6 if domain < 3 else 8 # set frame rate for dt=15 vs. 5 minutes
ani.save(filename_mp4, writer=animation.FFMpegWriter(fps=fps, bitrate=5000, codec="h264"), dpi=150)
/tmp/ipykernel_1683/1194218404.py:73: UserWarning: The input coordinates to pcolormesh are interpreted as cell centers, but are not monotonically increasing or decreasing. This may lead to incorrectly calculated cell edges, in which case, please supply explicit cell edges to pcolormesh.
pcm= ax.pcolormesh(lonslice.values, hamsl_raw.values, plotdata_raw.isel(Time=0).values,
(750, 149)
(15, 75)
(15, 75)
(15, 75)
(15, 75)
plotting frame 0
(15, 75)
(15, 75)
plotting frame 0
(15, 75)
(15, 75)
plotting frame 1
(15, 75)
(15, 75)
plotting frame 2
(15, 75)
(15, 75)
plotting frame 3
(15, 75)
(15, 75)
plotting frame 4
(15, 75)
(15, 75)
plotting frame 5
(15, 75)
(15, 75)
plotting frame 6
(15, 75)
(15, 75)
plotting frame 7
(15, 75)
(15, 75)
plotting frame 8
(15, 75)
(15, 75)
plotting frame 9
(15, 75)
(15, 75)
plotting frame 10
(15, 75)
(15, 75)
plotting frame 11
(15, 75)
(15, 75)
plotting frame 12
(15, 75)
(15, 75)
plotting frame 13
(15, 75)
(15, 75)
plotting frame 14
(15, 75)
(15, 75)
plotting frame 15
(15, 75)
(15, 75)
plotting frame 16
(15, 75)
(15, 75)
plotting frame 17
(15, 75)
(15, 75)
plotting frame 18
(15, 75)
(15, 75)
plotting frame 19
(15, 75)
(15, 75)
plotting frame 20
(15, 75)
(15, 75)
plotting frame 21
(15, 75)
(15, 75)
plotting frame 22
(15, 75)
(15, 75)
plotting frame 23
(15, 75)
(15, 75)
plotting frame 24
(15, 75)
(15, 75)
plotting frame 25
(15, 75)
(15, 75)
plotting frame 26
(15, 75)
(15, 75)
plotting frame 27
(15, 75)
(15, 75)
plotting frame 28
(15, 75)
(15, 75)
plotting frame 29
(15, 75)
(15, 75)
plotting frame 30
(15, 75)
(15, 75)
plotting frame 31
(15, 75)
(15, 75)
plotting frame 32
(15, 75)
(15, 75)
plotting frame 33
(15, 75)
(15, 75)
plotting frame 34
(15, 75)
(15, 75)
plotting frame 35
(15, 75)
(15, 75)
plotting frame 36
(15, 75)
(15, 75)
plotting frame 37
(15, 75)
(15, 75)
plotting frame 38
(15, 75)
(15, 75)
plotting frame 39
(15, 75)
(15, 75)
plotting frame 40
(15, 75)
(15, 75)
plotting frame 41
(15, 75)
(15, 75)
plotting frame 42
(15, 75)
(15, 75)
plotting frame 43
(15, 75)
(15, 75)
plotting frame 44
(15, 75)
(15, 75)
plotting frame 45
(15, 75)
(15, 75)
plotting frame 46
(15, 75)
(15, 75)
plotting frame 47
(15, 75)
(15, 75)
plotting frame 48
(15, 75)
(15, 75)
plotting frame 49
(15, 75)
(15, 75)
plotting frame 50
(15, 75)
(15, 75)
plotting frame 51
(15, 75)
(15, 75)
plotting frame 52
(15, 75)
(15, 75)
plotting frame 53
(15, 75)
(15, 75)
plotting frame 54
(15, 75)
(15, 75)
plotting frame 55
(15, 75)
(15, 75)
plotting frame 56
(15, 75)
(15, 75)
plotting frame 57
(15, 75)
(15, 75)
plotting frame 58
(15, 75)
(15, 75)
plotting frame 59
(15, 75)
(15, 75)
plotting frame 60
(15, 75)
(15, 75)
plotting frame 61
(15, 75)
(15, 75)
plotting frame 62
(15, 75)
(15, 75)
plotting frame 63
(15, 75)
(15, 75)
plotting frame 64
(15, 75)
(15, 75)
plotting frame 65
(15, 75)
(15, 75)
plotting frame 66
(15, 75)
(15, 75)
plotting frame 67
(15, 75)
(15, 75)
plotting frame 68
(15, 75)
(15, 75)
plotting frame 69
(15, 75)
(15, 75)
plotting frame 70
(15, 75)
(15, 75)
plotting frame 71
(15, 75)
(15, 75)
plotting frame 72
(15, 75)
(15, 75)
<Figure size 640x480 with 0 Axes>
QCloud + QIce for Domain 2, 29-Jan-2019, EDA09 Forcing:
corlasso_anim_qci_2019012900eda09d2_base.mp4
ds
<xarray.Dataset> Size: 59GB
Dimensions: (Time: 10, south_north: 865, west_east: 750,
bottom_top: 149)
Coordinates:
* Time (Time) datetime64[ns] 80B 2019-01-29T06:00:00 ... 2...
XLONG (south_north, west_east) float32 3MB dask.array<chunksize=(865, 750), meta=np.ndarray>
XLAT (south_north, west_east) float32 3MB dask.array<chunksize=(865, 750), meta=np.ndarray>
XTIME (Time) float32 40B dask.array<chunksize=(1,), meta=np.ndarray>
Dimensions without coordinates: south_north, west_east, bottom_top
Data variables: (12/52)
ITIMESTEP (Time) int32 40B dask.array<chunksize=(1,), meta=np.ndarray>
MUTOT (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
HGT (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
HAMSL (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
P_HYD (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
PRESSURE (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
... ...
MULFC (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
MULNB (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
MULPL (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
MUCAPE (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
MUCIN (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
REFL_10CM_MAX (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
Attributes: (12/39)
DX: 500.0
DY: 500.0
SIMULATION_START_DATE: 2019-01-29_06:00:00
WEST-EAST_GRID_DIMENSION: 751
SOUTH-NORTH_GRID_DIMENSION: 866
BOTTOM-TOP_GRID_DIMENSION: 150
... ...
doi_isPartOf_lasso-cacti: https://doi.org/10.5439/1905789
doi_isDocumentedBy: https://doi.org/10.2172/1905845
doi_thisFileType: https://doi.org/10.5439/1905819
history: processed by user d3m088 on machine cirrus89...
filename_user: corlasso_met_2019012900eda09d3_base_M1.m1.20...
filename_storage: corlassomet2019012900eda09d3baseM1.m1.201901...print(ds)
<xarray.Dataset> Size: 39GB
Dimensions: (Time: 10, bottom_top: 149, south_north: 865, west_east: 750)
Coordinates:
* Time (Time) datetime64[ns] 80B 2019-01-29T06:00:00 ... 2019-01-29...
XLONG (south_north, west_east) float32 3MB dask.array<chunksize=(865, 750), meta=np.ndarray>
XLAT (south_north, west_east) float32 3MB dask.array<chunksize=(865, 750), meta=np.ndarray>
XTIME (Time) float32 40B dask.array<chunksize=(1,), meta=np.ndarray>
Dimensions without coordinates: bottom_top, south_north, west_east
Data variables: (12/14)
ITIMESTEP (Time) int32 40B dask.array<chunksize=(1,), meta=np.ndarray>
QCLOUD (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
QRAIN (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
QICE (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
QSNOW (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
QGRAUP (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
... ...
REFL_10CM (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
CLDFRA (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
LWP (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
IWP (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
PRECIPWATER (Time, south_north, west_east) float32 26MB dask.array<chunksize=(1, 865, 750), meta=np.ndarray>
QNCLOUD (Time, bottom_top, south_north, west_east) float32 4GB dask.array<chunksize=(1, 50, 289, 250), meta=np.ndarray>
Attributes: (12/39)
DX: 500.0
DY: 500.0
SIMULATION_START_DATE: 2019-01-29_06:00:00
WEST-EAST_GRID_DIMENSION: 751
SOUTH-NORTH_GRID_DIMENSION: 866
BOTTOM-TOP_GRID_DIMENSION: 150
... ...
doi_isPartOf_lasso-cacti: https://doi.org/10.5439/1905789
doi_isDocumentedBy: https://doi.org/10.2172/1905845
doi_thisFileType: https://doi.org/10.5439/1905813
history: processed by user d3m088 on machine cirrus74...
filename_user: corlasso_cld_2019012900eda09d3_base_M1.m1.20...
filename_storage: corlassocld2019012900eda09d3baseM1.m1.201901...
print(theta.shape)
print(ds.THETA_E.shape)
print(ds2.QCLOUD.shape)
(865, 750)
(10, 149, 865, 750)
(10, 149, 865, 750)
print(lonslice.values.shape)
print(hamsl_raw.values.shape)
print(da_u.values.shape)
print(da_v.values.shape)
(750,)
(149, 750)
(10, 149, 750)
(10, 149, 750)
